Antarctic ecosystem
Climate
Krill (Euphausia superba)
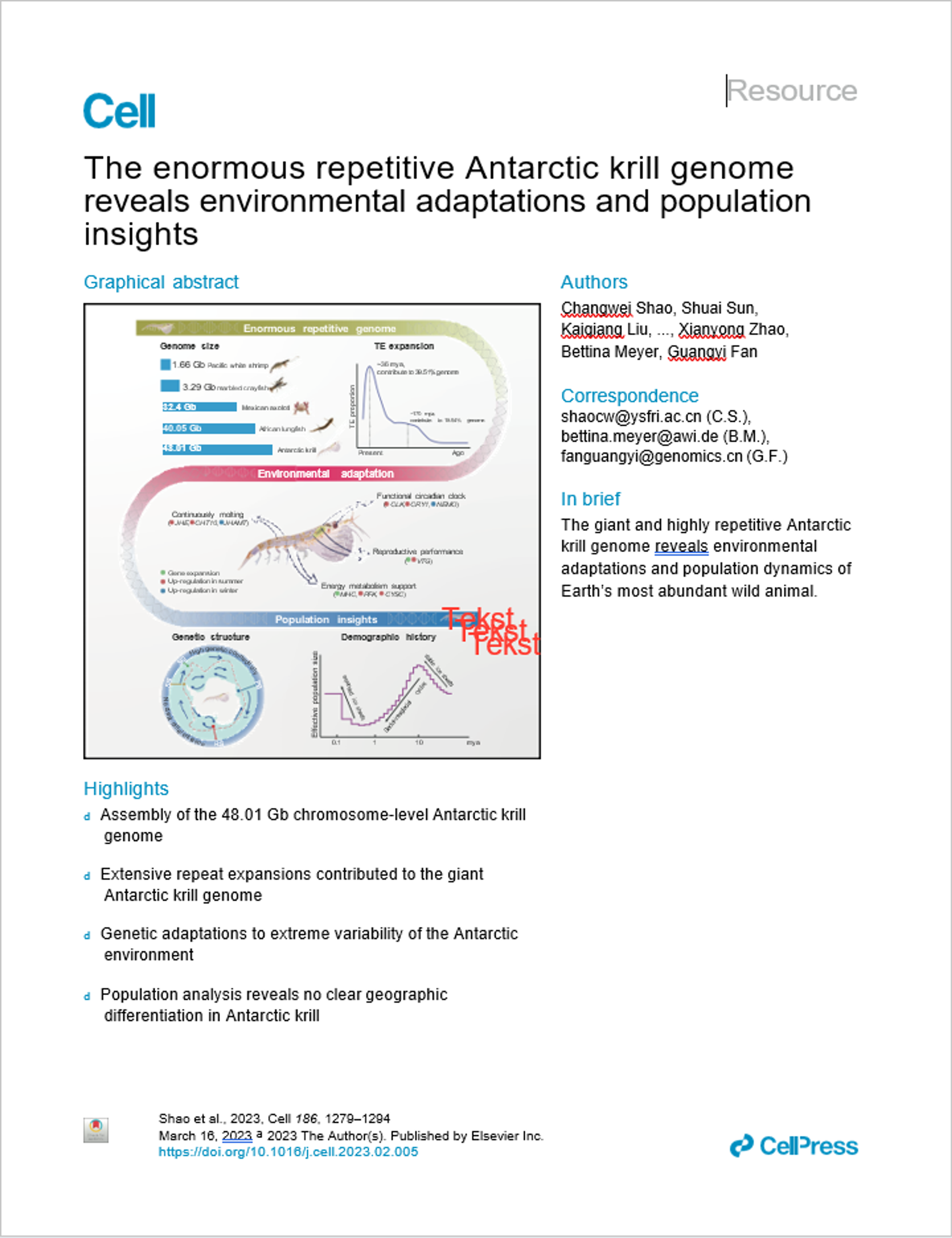
The Enormous Repetitive Antarctic Krill Genome
Summary
This groundbreaking study presents the first complete genome of Antarctic krill, revealing it to be the largest animal genome ever assembled at 48.01 gigabases - about 16 times larger than the human genome. The enormous size results from extensive repetitive DNA elements (72% of genome). Key findings include genetic adaptations for extreme Antarctic conditions through expanded gene families for continuous molting and energy metabolism, plus a functional circadian clock system. Population analysis of 75 individuals from four Antarctic regions shows high genetic connectivity with no major geographic barriers, but environmental selection is occurring. The genome provides crucial insights into how Earth's most abundant animal species has adapted to one of the planet's most extreme environments.
Key Findings
1
Genome Architecture: At 48.01 Gb, this represents the largest animal genome ever sequenced, with 72% composed of repetitive sequences, particularly DNA transposons 2
Environmental Adaptations: Identification of expanded gene families (25 significantly expanded) associated with continuous molting and energy metabolism, enabling survival in extreme seasonal Antarctic conditions 3
Circadian System: Complete molecular architecture of Antarctic krill circadian clock revealed, showing dual-feedback loop mechanism with seasonal expression differences 4
Population Structure: High genetic connectivity across Antarctica with no clear geographic differentiation, but evidence of environmental selection across different regions 5
Evolutionary History: Population bottleneck ~10 million years ago followed by expansion ~100,000 years ago, coinciding with major climate events